Bottom
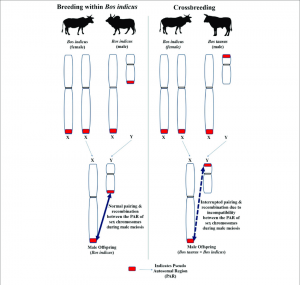
Cross localization during meiotic recombination of Bos taurus is mediated by the rapidly evolving zinc finger (ZF) domain of the PRDM9 gene. To study its impact on dairy cattle performance, we compared its genetic variation between relatively small Israeli (IL) Holsteins and North American (USA) Holsteins numbering in the millions.
Discussion
The purpose of this study was to discover PRDM9 alleles that affect milk production characteristics or could lead to faster genomic selection in Holstein breeding cattle by controlling meiotic recombination rates. On the one hand, PRDM9-driven meiotic recombination can induce deleterious chromosomal instability and meiotic drive; on the other, it rearranges paternal and maternal genetic alleles in the next generation, which could provide better novel combinations of genetic variants.
A recent study of US cattle showed that a specific PRDM9 allele termed “allele 5” has a dramatic influence on the location of recombination hotspots and unique recombination hotspot regions that are distinguished from hotspot regions modulated by all other alleles.
However, it was unclear if this pattern would also be seen in IL Holstein cattle, which have a different demographic history, although AI is often practised with semen from elite US bulls to improve the local variety. We applied genome sequencing as the putative method of choice to investigate which PRDM9 alleles are prevalent in Holstein ILs.
Computer cloning of the PRDM9 gene from an influential IL Holstein bull indicated that it encodes a 727 amino acid variant PRDM9, which we refer to as the dairy form. This form was longer than the bovine form of the GenBank reference sequence, which was derived from the Hereford cattle breed, as the genome of this breed was the first to be sequenced, assembled, and annotated.
Furthermore, this reference suggests alternative splicing that does not correspond to the common transcript, as demonstrated by our assembly of RNA-Seq data obtained from Hereford SuperBull 99,375 testes; as such, it should be considered a computational artefact. Based on this RNA-Seq readout assembly, we provide the correct transcription sequence of the meat form.
Results
Initially, we analyzed the major BTA1 haplotypes present in IL Holsteins based on the 10 most telomeric SNPs of the BovineSNP50 BeadChip. Sequencing of representative haplotype carriers indicated that for all common haplotypes (> 6%), the variable PRDM9 ZnF array consisted of seven tandem ZF repeats. Two rare haplotypes (frequency < 4%) carried a PRDM9 indication, while all others were taurine-like variants.
These two haplotypes included the minor SNP allele, which was closely related to a previously described PRDM9 allele known to induce the unique localization of recombination hotspots. One of them had a significant negative effect (p = 0.03) on the fertility of the IL bull. This haplotype combined the rare minor alleles of the single SNPs with significant (p < 0.05) negative substitution effects on fertility of US sires (SCR).
Analysis of telomeric SNPs indicated overall agreement of allele frequencies (R = 0.95) and substitution effects on bull fertility (SCR, R = 0.6) between the US and UK samples. IL. Surprisingly, alleles that had a negative impact on male fertility had the most positive substitution effects on female fertility traits (DPR, CCR, and HCR).
Conclusions
A negative genetic correlation between male and female fertility is encoded within the BTA1 telomere. By cloning the PRDM9 taurine gene, which is the common form Holsteins carry, we found infiltration of a PRDM9 variant in this population. During meiosis, in heterozygous males, the PRDM9 signalling variant can induce recombination breakpoint incompatibility and male infertility.
However, this variant is associated with favourable female fertility, which would explain its survival and the overall negative correlation (R = − 0.3) observed between male and female fertility in American Holsteins. More research is needed to explain the mechanism behind this positive effect and to devise a methodology to disassociate it from the negative effect on male fertility during reproduction.